Multiplex Ligation Probe Amplification or MLPA for short. The MLPA technique was first developed by MRC Holland. It's a technique which can be used to identify small copy number changes of DNA or RNA sequences, i.e., whether there is a deletion or duplication of a specific region of the genome. It can also be used to determine the methylation status of imprinted or promoter regions, and it can also be used to detect known points mutations, and single nucleotide polymorphisms, or SNPs. We are going to consider each of these differences in turn in this presentation. But first, we're going to think about how MLPA works. There are five basic steps that take place during the MLPA process.
The first is DNA denaturation, or separation, of the target strands of DNA. The second is hybridisation. Hybridisation of a specific MLPA probe to a specific region of the genome. The third is ligation, or both ligation and digestion, depending on what type of MLPA you are undertaking. The fourth is amplification of the probe. And the fifth is capillary electrophoresis to separate the products of the amplification process for analysis. Let's now discuss how MLPA works. MLPA probes consist of a pair of oligonucleotides which recognise adjacent target sites on the patient's DNA. The first oligonucleotide contains a region complementary to a forward primer. And the second oligonucleotide contains a region complementary to the reverse primer.
The second oligonucleotide also contains a stuffer sequence which can be of varying lengths. Denaturation of the patient DNA causes it to become single stranded, so that the oligonucleotides can hybridise to their target DNA.
 In this figure, you can see two MLPA probes. One for Target A and one for Target B. And each of them consist of paired oligonucleotides. You can note that the stuffer sequence is a different length in each probe. Both oligonucleotides in this pair have correctly hybridised to their adjacent target sequence. This allows the next step of the process, ligation, to take place. If both probe target sequences have hybridised correctly and adjacently, they can be ligated together by a thermostable ligase.So now we have a ligated probe which has joined our paired oligonucleotides together. You will remember that the first oligonucleotide contained a region complementary to a forward primer, and the second oligonucleotide contained a region complementary to the reverse primer. Now that these oligonucleotides have been ligated together, the addition of an amplification step, using the forward and reverse primers, allows the full length of the probe to be amplified. Due to the presence of the stuffer sequence of varying length, the amplification product of each probe will be of a unique length.The fact that each probe will lead to the creation of amplification products of a specific and different length to the other probes means that the amplification products can now be separated by electrophoresis. And each individual amplification product will give information about a single probe within the mixture. The relative amounts of the probe amplification product will reflect the relative copy number of the target sequences.
In this figure, you can see two MLPA probes. One for Target A and one for Target B. And each of them consist of paired oligonucleotides. You can note that the stuffer sequence is a different length in each probe. Both oligonucleotides in this pair have correctly hybridised to their adjacent target sequence. This allows the next step of the process, ligation, to take place. If both probe target sequences have hybridised correctly and adjacently, they can be ligated together by a thermostable ligase.So now we have a ligated probe which has joined our paired oligonucleotides together. You will remember that the first oligonucleotide contained a region complementary to a forward primer, and the second oligonucleotide contained a region complementary to the reverse primer. Now that these oligonucleotides have been ligated together, the addition of an amplification step, using the forward and reverse primers, allows the full length of the probe to be amplified. Due to the presence of the stuffer sequence of varying length, the amplification product of each probe will be of a unique length.The fact that each probe will lead to the creation of amplification products of a specific and different length to the other probes means that the amplification products can now be separated by electrophoresis. And each individual amplification product will give information about a single probe within the mixture. The relative amounts of the probe amplification product will reflect the relative copy number of the target sequences.
Here's an example of what an MLPA trace would look like once the amplification probes have been separated. Here, MLPA has been undertaken for a gene called the APC gene. Each of these blue peaks represents a different probe that was targeted to a specific region of the APC gene.
You can see that the relative height of these blue lines indicate the amount of amplification product that was available for that specific probe. So if you compare the control sample to the patient sample, you can see that for Probes 11, 12, and 13, there is a half reduction in height in the case compared to the control. This indicates that there was half the amount of amplification product available, and therefore, that there was a heterozygous deletion of Exons 11 to 13 in this patient sample.
In the slide, we can see that the differences in the relative amounts of case and control DNA are reflected in the different relative peaks of the amplified probes for that region, and that can indicate if there was a deletion or a duplication in the patient DNA in that region. As well as detecting deletions and duplications, MLPA can also be used to undertake methylation-specific analysis of an imprinted region. The first three steps of MS MLPA are the same as MLPA. But a ligation and digestion step is now included.
In this figure, we see two targets. The target on the left is methylated, and the target on the right is unmethylated.The DNA is first denatured, as it was previously, splitting it into single strands. The MLPA oligo probes then hybridise to the target sequence. Simultaneous ligation and digestion of the probe DNA complex then occurs. The digestion uses a methylation-specific endonuclease. This only targets unmethylated DNA. Therefore, if the DNA is methylated, it will be protected from the endonuclease, and a normal MLPA product will be detected. This is shown on the left-hand side of the slide. If it is not methylated, the probe product will be digested, and no amplification product can be formed. Therefore, by looking for the presence, or not, of probe, we can tell whether that methylation target was methylated or not.
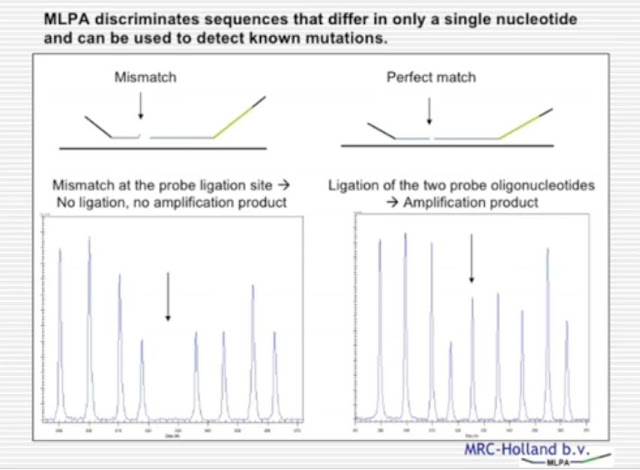
Finally, MLPA can be used for the detection of point mutations. This is based upon the principle that the oligonucleotides are exquisitely sensitive to the target DNA sequence they are designed to hybridise to. If there is a SNP or a mutation below the probe hybridisation site, then the probes will not hybridise, and no amplification product will be produced. Therefore, if there is a common mutation in a gene, a probe can be designed specific to this mutation, so the signal will only be produced when the common mutation, or SNP, is present.
This is shown in the diagram here. The diagram on the left shows normal DNA without an associated signal.
This is because the oligonucleotides have not hybridised, could not be ligated, and therefore, the probe could not be amplified. On the right; however, there's a common mutation. This allows both oligonucleotides to hybridise, ligation to occur, and an amplification product to be produced.
In summary, there are three main clinical applications for MLPA. It can be used to identify exon deletions or duplications within single genes. It can also be used to identify microdeletion syndromes, such as Di George Syndrome. Secondly, it is a powerful and rapid technique to detect methylation abnormalities. And finally, it can be used to identify common mutations.
The first is DNA denaturation, or separation, of the target strands of DNA. The second is hybridisation. Hybridisation of a specific MLPA probe to a specific region of the genome. The third is ligation, or both ligation and digestion, depending on what type of MLPA you are undertaking. The fourth is amplification of the probe. And the fifth is capillary electrophoresis to separate the products of the amplification process for analysis. Let's now discuss how MLPA works. MLPA probes consist of a pair of oligonucleotides which recognise adjacent target sites on the patient's DNA. The first oligonucleotide contains a region complementary to a forward primer. And the second oligonucleotide contains a region complementary to the reverse primer.
The second oligonucleotide also contains a stuffer sequence which can be of varying lengths. Denaturation of the patient DNA causes it to become single stranded, so that the oligonucleotides can hybridise to their target DNA.
 In this figure, you can see two MLPA probes. One for Target A and one for Target B. And each of them consist of paired oligonucleotides. You can note that the stuffer sequence is a different length in each probe. Both oligonucleotides in this pair have correctly hybridised to their adjacent target sequence. This allows the next step of the process, ligation, to take place. If both probe target sequences have hybridised correctly and adjacently, they can be ligated together by a thermostable ligase.So now we have a ligated probe which has joined our paired oligonucleotides together. You will remember that the first oligonucleotide contained a region complementary to a forward primer, and the second oligonucleotide contained a region complementary to the reverse primer. Now that these oligonucleotides have been ligated together, the addition of an amplification step, using the forward and reverse primers, allows the full length of the probe to be amplified. Due to the presence of the stuffer sequence of varying length, the amplification product of each probe will be of a unique length.The fact that each probe will lead to the creation of amplification products of a specific and different length to the other probes means that the amplification products can now be separated by electrophoresis. And each individual amplification product will give information about a single probe within the mixture. The relative amounts of the probe amplification product will reflect the relative copy number of the target sequences.
In this figure, you can see two MLPA probes. One for Target A and one for Target B. And each of them consist of paired oligonucleotides. You can note that the stuffer sequence is a different length in each probe. Both oligonucleotides in this pair have correctly hybridised to their adjacent target sequence. This allows the next step of the process, ligation, to take place. If both probe target sequences have hybridised correctly and adjacently, they can be ligated together by a thermostable ligase.So now we have a ligated probe which has joined our paired oligonucleotides together. You will remember that the first oligonucleotide contained a region complementary to a forward primer, and the second oligonucleotide contained a region complementary to the reverse primer. Now that these oligonucleotides have been ligated together, the addition of an amplification step, using the forward and reverse primers, allows the full length of the probe to be amplified. Due to the presence of the stuffer sequence of varying length, the amplification product of each probe will be of a unique length.The fact that each probe will lead to the creation of amplification products of a specific and different length to the other probes means that the amplification products can now be separated by electrophoresis. And each individual amplification product will give information about a single probe within the mixture. The relative amounts of the probe amplification product will reflect the relative copy number of the target sequences.Here's an example of what an MLPA trace would look like once the amplification probes have been separated. Here, MLPA has been undertaken for a gene called the APC gene. Each of these blue peaks represents a different probe that was targeted to a specific region of the APC gene.
You can see that the relative height of these blue lines indicate the amount of amplification product that was available for that specific probe. So if you compare the control sample to the patient sample, you can see that for Probes 11, 12, and 13, there is a half reduction in height in the case compared to the control. This indicates that there was half the amount of amplification product available, and therefore, that there was a heterozygous deletion of Exons 11 to 13 in this patient sample.
In the slide, we can see that the differences in the relative amounts of case and control DNA are reflected in the different relative peaks of the amplified probes for that region, and that can indicate if there was a deletion or a duplication in the patient DNA in that region. As well as detecting deletions and duplications, MLPA can also be used to undertake methylation-specific analysis of an imprinted region. The first three steps of MS MLPA are the same as MLPA. But a ligation and digestion step is now included.
In this figure, we see two targets. The target on the left is methylated, and the target on the right is unmethylated.The DNA is first denatured, as it was previously, splitting it into single strands. The MLPA oligo probes then hybridise to the target sequence. Simultaneous ligation and digestion of the probe DNA complex then occurs. The digestion uses a methylation-specific endonuclease. This only targets unmethylated DNA. Therefore, if the DNA is methylated, it will be protected from the endonuclease, and a normal MLPA product will be detected. This is shown on the left-hand side of the slide. If it is not methylated, the probe product will be digested, and no amplification product can be formed. Therefore, by looking for the presence, or not, of probe, we can tell whether that methylation target was methylated or not.
Finally, MLPA can be used for the detection of point mutations. This is based upon the principle that the oligonucleotides are exquisitely sensitive to the target DNA sequence they are designed to hybridise to. If there is a SNP or a mutation below the probe hybridisation site, then the probes will not hybridise, and no amplification product will be produced. Therefore, if there is a common mutation in a gene, a probe can be designed specific to this mutation, so the signal will only be produced when the common mutation, or SNP, is present.
This is shown in the diagram here. The diagram on the left shows normal DNA without an associated signal.
This is because the oligonucleotides have not hybridised, could not be ligated, and therefore, the probe could not be amplified. On the right; however, there's a common mutation. This allows both oligonucleotides to hybridise, ligation to occur, and an amplification product to be produced.
In summary, there are three main clinical applications for MLPA. It can be used to identify exon deletions or duplications within single genes. It can also be used to identify microdeletion syndromes, such as Di George Syndrome. Secondly, it is a powerful and rapid technique to detect methylation abnormalities. And finally, it can be used to identify common mutations.
Source:- https://www.futurelearn.com/courses/molecular-techniques/6/steps/765686







No comments:
Post a Comment